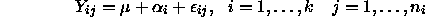
Last time we also talked about the ``grand mean plus effects'' parameterization,
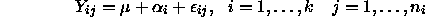
The relationship of this model to the cell means model

is that 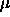 is supposed to be an average of the
is supposed to be an average of the 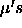 , and
that
, and
that 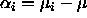 .
.
The grand mean plus effects model has k+1 parameters, 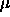 ,
,
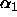 , ...,
, ..., 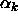 and only k cell means to estimate, so
some sort of linear constraint is needed to make the model work.
There are several different ways to impose a ``sum to zero''
constraint, each corresponding to a different ``grand mean'':
and only k cell means to estimate, so
some sort of linear constraint is needed to make the model work.
There are several different ways to impose a ``sum to zero''
constraint, each corresponding to a different ``grand mean'':
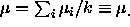 , it follows that
, it follows that
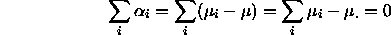
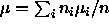 , then it follows that
, then it follows that
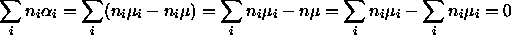
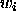 such that
such that 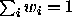 and we define
and we define  , it
similarly follows that
, it
similarly follows that


is an unbiased estimator of the grand mean 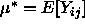 . It
can be shown that if
. It
can be shown that if 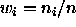 (sample size weights) this estimator
has the least possible variance (this is a nice exercise in
multivariate differential calculus).
(sample size weights) this estimator
has the least possible variance (this is a nice exercise in
multivariate differential calculus).
It's easy to see that the model.tables() function gives you the sample-size weighted effects when you specify type="effects":
402 > model.tables(coag.aov,type="effects")
Refitting model to allow projection
Tables of effects
diet
A B C D
-1.875 2.833 2.5 -4.25
rep 8.000 6.000 6.0 4.00
402 > sum(scan())
1: -1.875 2.833 2.5 -4.25
5:
[1] -0.792
402 > sum(scan()*scan())
1: -1.875 2.833 2.5 -4.25
5:
1: 8.000 6.000 6.0 4.00
5:
[1] -0.002
One can fit the unweighted effects model directly by telling SPLUS what kind of contrasts to use:
402 > coag.sum.aov _ aov(coag ~ diet,data=coag,contrasts=list(diet=contr.sum))
402 > coefficients(coag.sum.aov)
(Intercept) diet1 diet2 diet3
63.80208 -1.677083 3.03125 2.697917
402 > co _ .Last.value
402 > c(co[2:4],-sum(co[2:4]))
diet1 diet2 diet3
-1.677083 3.03125 2.697917 -4.052083
402 > co[1] + c(co[2:4],-sum(co[2:4]))
diet1 diet2 diet3
62.125 66.83333 66.5 59.75
402 > model.tables(coag.aov,type="means")$tables$diet
Refitting model to allow projection
A B C D
62.125 66.83333 66.5 59.75
It is also possible to use summary.lm to look at the individual
coefficients and their standard errors.
402 > summary.lm(coag.sum.aov)
Call: aov(formula = coag ~ diet, data = coag, contrasts = list(diet = contr.sum))
Residuals:
Min 1Q Median 3Q Max
-3.833 -2.583 0.375 1.312 4.5
Coefficients:
Value Std. Error t value Pr(>|t|)
(Intercept) 63.8021 0.5838 109.2923 0.0000
diet1 -1.6771 0.9066 -1.8499 0.0792
diet2 3.0313 0.9911 3.0585 0.0062
diet3 2.6979 0.9911 2.7221 0.0131
Residual standard error: 2.775 on 20 degrees of freedom
Multiple R-Squared: 0.5472
F-statistic: 8.056 on 3 and 20 degrees of freedom, the p-value is 0.001028
Correlation of Coefficients:
(Intercept) diet1 diet2
diet1 -0.1894
diet2 -0.0346 -0.2454
diet3 -0.0346 -0.2454 -0.3061
402 > #
402 > # now use the information in the summary to construct a
402 > # point estimate and se for the effect of the fourth diet ("D")
402 > #
402 > diet4 _ c(0,-1,-1,-1)
402 > diet4 %*% coefficients(coag.sum.aov)
[,1]
[1,] -4.052083
402 > diet4 %*% summary.lm(coag.sum.aov)$cov.unscaled %*% diet4
[,1]
[1,] 0.1692708
402 > sqrt(.Last.value)
[,1]
[1,] 0.4114254
One can do something similar with the cell means model:
402 > coag.tx.aov _ aov(coag ~ diet,data=coag,contrasts=list(diet=contr.treatment))
402 > summary.lm(coag.tx.aov)
Call: aov(formula = coag ~ diet, data = coag, contrasts = list(diet = contr.treatmen
t))
Residuals:
Min 1Q Median 3Q Max
-3.833 -2.583 0.375 1.313 4.5
Coefficients:
Value Std. Error t value Pr(>|t|)
(Intercept) 62.1250 0.9809 63.3322 0.0000
dietB 4.7083 1.4984 3.1422 0.0051
dietC 4.3750 1.4984 2.9198 0.0085
dietD -2.3750 1.6990 -1.3979 0.1775
Residual standard error: 2.775 on 20 degrees of freedom
Multiple R-Squared: 0.5472
F-statistic: 8.056 on 3 and 20 degrees of freedom, the p-value is 0.001028
Correlation of Coefficients:
(Intercept) dietB dietC
dietB -0.6547
dietC -0.6547 0.4286
dietD -0.5774 0.3780 0.3780
Now SPLUS has parametrized the model so that the intercept is the cell
mean for Diet A, and the other parameters are the offsets from Diet A
to Diets B, C, and D.
SPLUS has several kinds of built-in ``contrast'' options, that are used to reparametrize an ANOVA model. They are:
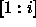 , in
the unweighted effects model. So for example if
, in
the unweighted effects model. So for example if 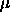 is the grand
mean and
is the grand
mean and 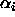 are the unweighted effects for level i (so all
the
are the unweighted effects for level i (so all
the 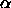 's sum to zero), the parameters
's sum to zero), the parameters 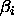 for the Helmert
contrasts are related to the effects
for the Helmert
contrasts are related to the effects 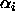 in the unweighted
effects model by
in the unweighted
effects model by
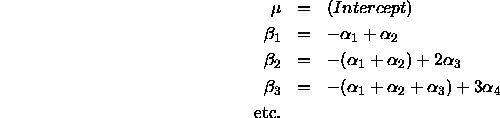
[to see this, compare model.matrix(coag.sum.aov)[1:4,] with model.matrix(coag.aov)[1:4,] above!]