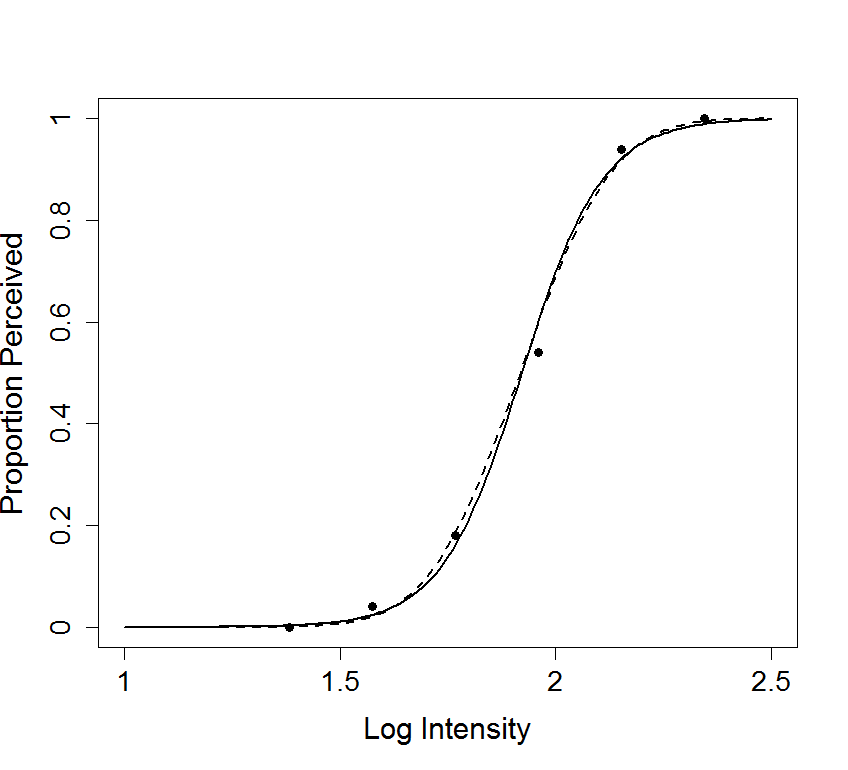
# Figure 14.1 - Plots actual data, a logistic regression, and a
# probit regression, in order to show that (a) the fits are good, and
# (b) the logistic and probit models are very similar.
#
# Figure caption: Two curves fitted to the data in Figure 8.9. The
# fitted curve from probit regression (dashed line) is shown together
# with the fitted curve from logistic regression. The fits are very
# close to eachother.
# MPDiR loads the HSP data set. The data are from Hecht et al, and
# show, in 50 trials, whether or not light flashes of a certain log10
# intensity were observed by a subject.
#
library(MPDiR)
chosenRun <- "R1" # Use Run 1.
chosenSubject <- "SS" # Choose the subject S.S.
chosenData <- HSP$Run == chosenRun & HSP&Obs == chosenSubject
dataForOneRun <- HSP[chosenData, ]
# Pull the data for one run.
lightIntensiy <- log10(dataForOneRun$Q)
percentDetected <- dataForOneRun$p / 100 # Scale percentages to (0,1).
# Fit logistic and probit models.
modelEquation <- percentDetected ~ lightIntensity
logisticModel <- glm(modelEquation, family = binomial(link = "logit"))
probitModel <- glm(modelEquation, family = binomial(link = "probit"))
lightValues <- data.frame( x = seq(from = 1, to = 2.5, by = 0.01))
# Plot actual points and the models' fits.
plot(lightIntensity, percentDetected)
xlim = c(1, 2.5), ylim = c(0, 1),
xlab = "Log Intensity", ylab = "Proportion Perceived", main = "",
xaxt = "n", yaxt = "n", pch = 16, cex.lab = 1.5)
lines(lightValues, predict(logisticModel, newdata = lightValues),
lty = 1, lwd = 2)
lines(lightValues, predict(probitModel, newdata = lightValues),
lty = 2, lwd = 2)
axis(1, at = seq(from = 1, to = 2.5, by = 0.5), cex.axis = 1.4)
axis(2, at = seq(from = 0, to = 1, by = 0.2), cex.axis = 1.4)
#legend("topleft", c("Logistic", "Probit"), lty = c(1, 2),
# lwd = c(2, 2), cex = 1.5)
dev.print(device = postscript, "14.1.eps", horizontal = TRUE)