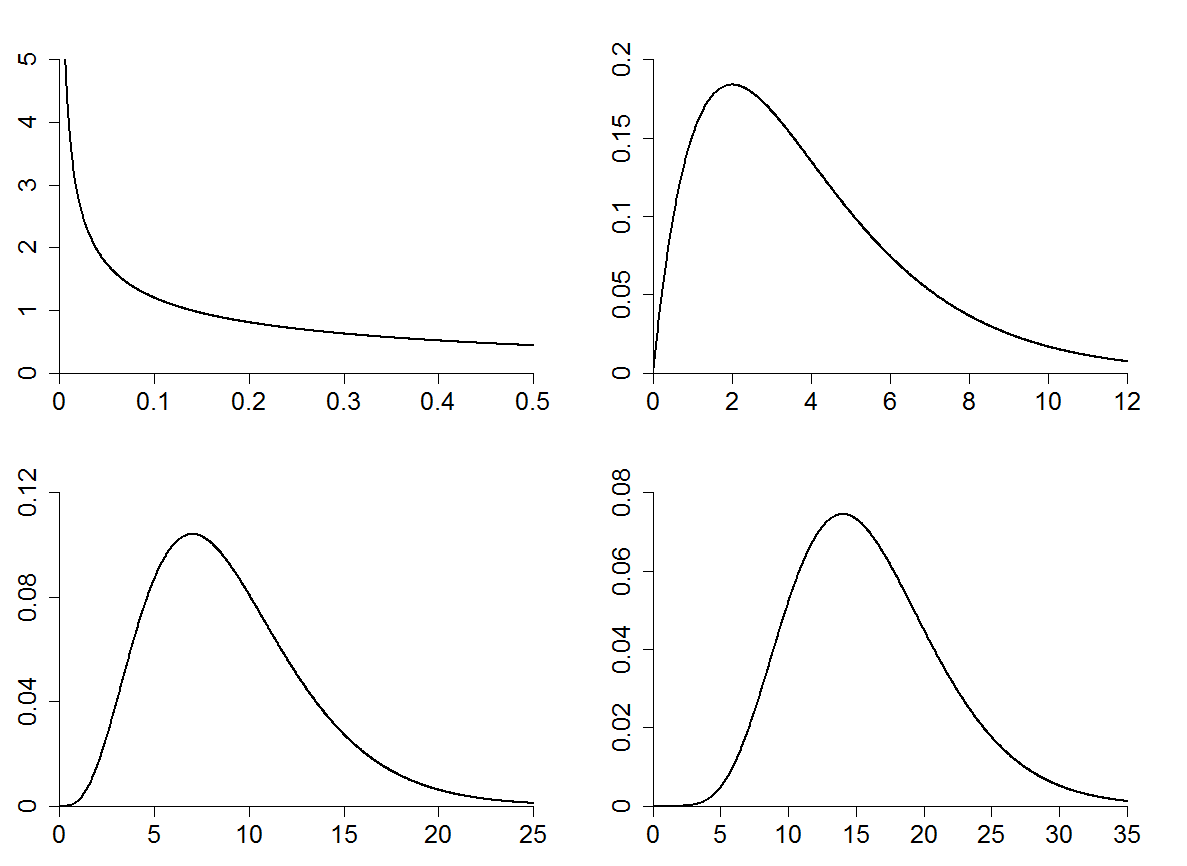
# This script plots four chi-squared densities, with 1, 4, 9, and 16
# degrees of freedom, respectively.
#
# We first set up the varying elements (the distribution parameters,
# and the limits and tick-marks on the x-axes), and then loop through
# to plot each subplot with the same style.
# Set up the graphics device.
par(mfrow = c(2, 2))
par(xaxs = "i")
par(yaxs = "i")
par(mar = rep(3, 4))
# Set some graphical parameters.
line.type <- 1
line.width <- 2
# Set the changing parameters for each plot.
degreesOfFreedomList <- c(1, 4, 9, 16)
xMaxima <- c(0.5, 12, 25, 35)
yMaxima <- c(5, 0.2, 0.12, 0.08)
xTickMarks <- c(0.1, 2, 5, 5)
yTickMarks <- c(1, 0.05, 0.04, 0.02)
resolution <- 1000
axis.size <- 1.5
# Loop through and plot.
for (i in 1:4) {
xMax <- xMaxima[i]
yMax <- yMaxima[i]
xTickMark <- xTickMarks[i]
yTickMark <- yTickMarks[i]
degreesOfFreedom <- degreesOfFreedomList[i]
x.values <- seq(from = 0, to = xMax, length.out = resolution)
chi.squared.pdf.values <- dchisq(x.values, df = degreesOfFreedom)
plot(x.values, chi.squared.pdf.values,
xlim = c(0, xMax), ylim = c(0, yMax),
type = "l", lty = line.type, lwd = line.width,
main = "", xlab = "", ylab = "",
bty = "n", xaxt = "n", yaxt = "n")
x.axis <- seq(from = 0, to = xMax, by = xTickMark)
y.axis <- seq(from = 0, to = yMax, by = yTickMark)
axis(1, at = x.axis, labels = x.axis, cex.axis = axis.size)
axis(2, at = y.axis, labels = y.axis, cex.axis = axis.size)
}
# Close the graphics device.
dev.print(device = postscript, "5.4.eps", horizontal = TRUE)