The file geyser.dat contains data about Old Faithful Geyser,
a popular tourist attraction. A geologist was interested in predicting the
length of time between eruptions based on the duration of eruptions. He
observed the geyser several times in August 1978, and several more times
in August 1979.
> geyser <- read.table("geyser.dat",col.names=c("date","interval","duration","is.aug.78","is.aug.79"))
> attach(geyser)
The data file consists of what day the observation took place, the interval
between eruptions (in minutes), the duration of the preceding eruption (in
minutes), a 0/1 indicator variable for August 1978, and another 0/1 indicator
variable for August 1979.
> motif()
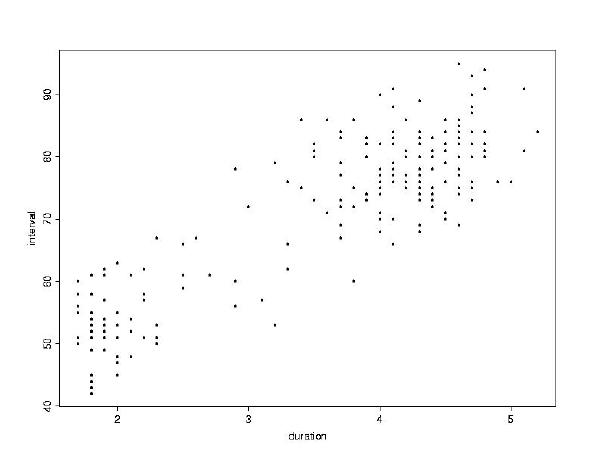
> plot(duration, interval)

duration
and interval. The longer the eruption, the more time until
the next eruption.
Let's start with only one predictor, duration. The formula
would be ``interval modeled by duration'', which is interval ~ duration
. The function to create a regression model is lm, for Linear
Model.
> lm(interval ~ duration)The
Call:
lm(formula = interval ~ duration)
Coefficients:
(Intercept) duration
33.90084 10.37363
Degrees of freedom: 221 total; 219 residual
Residual standard error: 6.170498
lm function creates an object of class ``linear model'',
and returns its value. The value includes the coefficients and other information.
It is a good idea to store this object someplace so that you can update
it and refer to it as you proceed with the analysis.
> geyser.lm <- lm(interval ~ duration)The
summary function, which you have probably used to generate
summary statistics about a vector, comes in handy with model objects.
> summary(geyser.lm)The
Call: lm(formula = interval ~ duration)
Residuals:
Min 1Q Median 3Q Max
-14.1 -4.507 -0.4701 4.194 16.83
Coefficients:
Value Std. Error t value Pr(>|t|)
(Intercept) 33.9008 1.4409 23.5277 0.0000
duration 10.3736 0.3850 26.9428 0.0000
Residual standard error: 6.17 on 219 degrees of freedom
Multiple R-Squared: 0.7682
F-statistic: 725.9 on 1 and 219 degrees of freedom, the p-value is 0
Correlation of Coefficients:
(Intercept)
duration -0.9576
summary command provides a bevy of useful information, including
p-values, multiple R-Squared, and correlation. Based on this information,
it looks like duration is a very good predictor of interval
.
Another way to examine regression output is graphically, using the
plot command. When given regression output as its argument,
plot will produce six diagnostic plots. To see them all, you must
split the plotting window into six or more parts. For instance, make it into
a 2 by 3 grid of windows.
> par(mfrow=c(2,3))
> plot(geyser.lm)
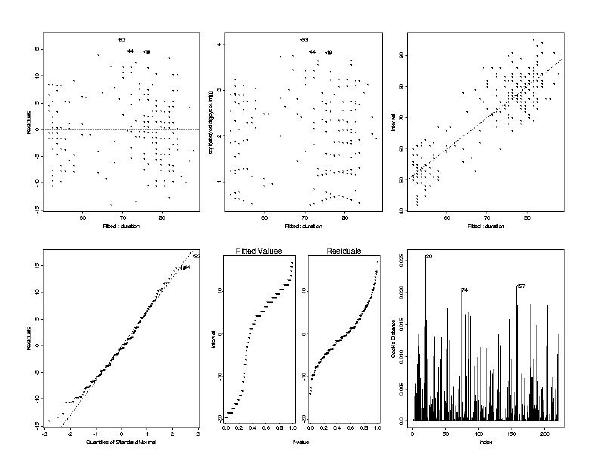
par(mfrow=c(2,3)) command divided the graphics window into
six separate windows. To change it back to normal, do par(mfrow=c(1,1))
(that is, one row by one column). This command will take effect when the
next plot is executed.
The six plots show useful diagnostic information, such as plots of residuals vs. fitted values, a quantile-normal plot, and a Cook's distance plot (the influence of each data point on the final model, large values indicate high influence).
Other functions which can be used on model objects include residuals
(returns a vector of the residuals) and fitted (returns a vector
of the fitted values).